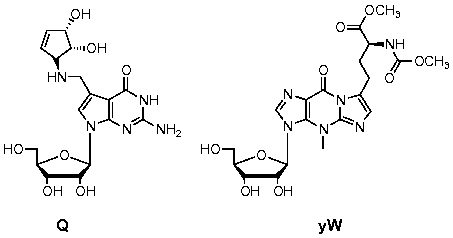
The Zhang Lab is dedicated to studying the role of RNA modifications, the next scientific frontier. There is emerging evidence that RNA modifications are functionally significant and play important roles in biological processes and diseases in vertebrates.
Through our research, we aim to:
- Probe the wide spectrum of structure and biological function of RNA modifications
- Understand how RNA modifications interact with one another to deliver a scope of broad yet controlled biological function and regulation
- Quantitatively understand how the level of RNA modifications change in response to changes in cellular environment and environmental stress
Successful achievement of these goals will represent not only a significant progression in our understanding towards RNA epigenetics and nucleic acid biochemistry but also a solid step towards the logical development of targeted therapy against diseases resultant from aberrant RNA modification patterns.
The Zheng Lab is affiliated with the University at Albany’s Department of Chemistry and RNA Institute.
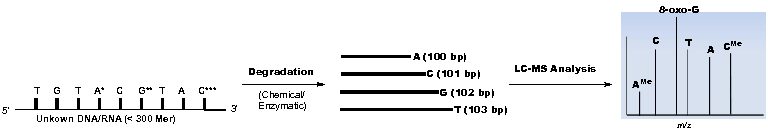
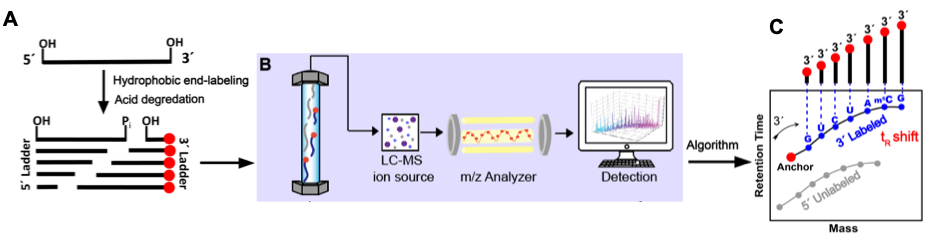
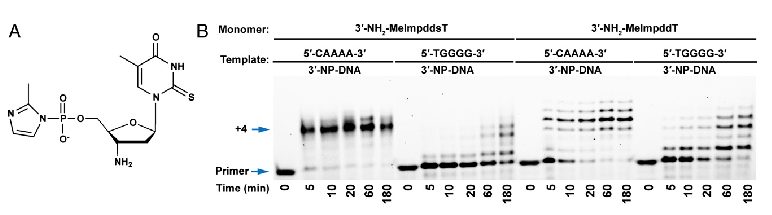
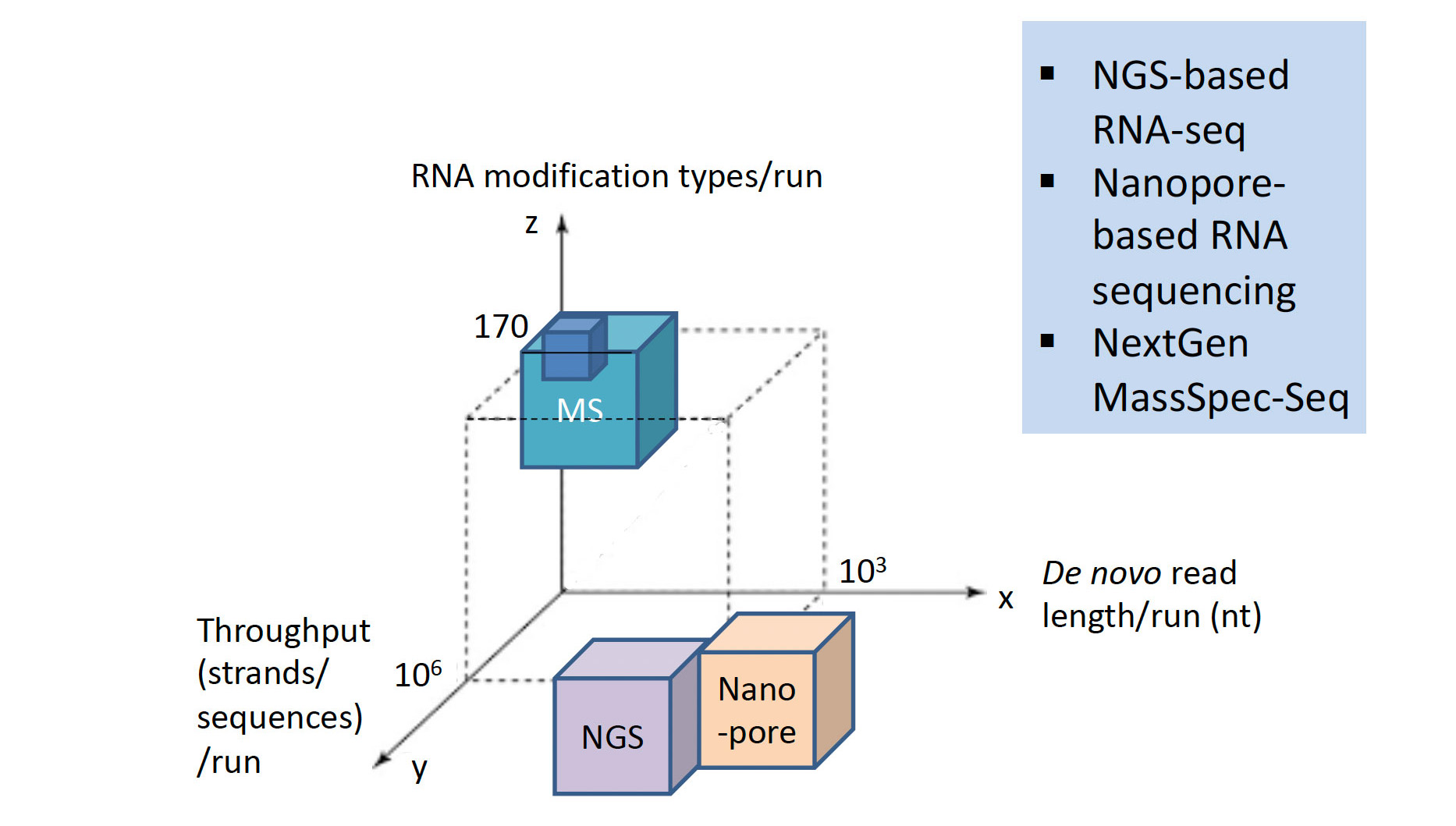
Major RNA Sequencing Platform Technologies
Join the Zhang Lab
We appreciate your interest in joining the Zhang Lab. Potential projects for new lab members include, but not limited to, the following:
- Methodological development for the direct sequencing of RNAs and their modifications
- Development of facile and robust strategies for the synthetic preparation of non-canonical ribonucleosides and their incorporations into RNA
- Probing the breadth of regulatory roles of RNA modifications in gene expression
- Systems-level mapping of the roles of RNA modifications, and the modifications’ interactions with other cellular components, into a holistic regulatory network
For more information, please contact Dr. Zhang directly at [email protected].
NIH-funded Postdoc (Position Available Immediately)
Professor Shenglong Zhang’s laboratory, which is affiliated with the University at Albany’s Department of Chemistry and RNA Institute, seeks a research scientist, technician or equivalent for development of next generation mass spectrometry-based RNA sequencing methods.
This includes development of methods to directly sequence RNAs, to examine their associated modifications, and to explore the potential roles of RNA modifications, e.g., in COVID-19, cancer biology, and metabolic diseases.
The candidate will work for an NIH-funded interdisciplinary research project, and this position is renewable annually contingent upon the continuous NIH support to the project.
Candidates will have ample opportunities to acquire and develop new skills, work closely in a supportive, highly collaborative and energetic environment with the PIs and collaborators in Columbia University and DirectSeq Biosciences, Inc., and communicate results to the scientific community through conference presentations, patents and peer-reviewed publications.
Qualifications:
- A PhD (or equivalent), with strong backgrounds in nucleic acid chemistry and RNA biology
- Knowledge in sequencing technique, liquid chromatography and mass spectrometric methods is strongly preferred
- Experience in computer programming using Python/Java, and/or algorithmic development is a plus
The position is open immediately although the start date is flexible.
To apply, please send your curriculum vitae, a description of previous research, research accomplishments, career interests and three references to Dr. Zhang at [email protected].
Applications will be considered until the position is filled.
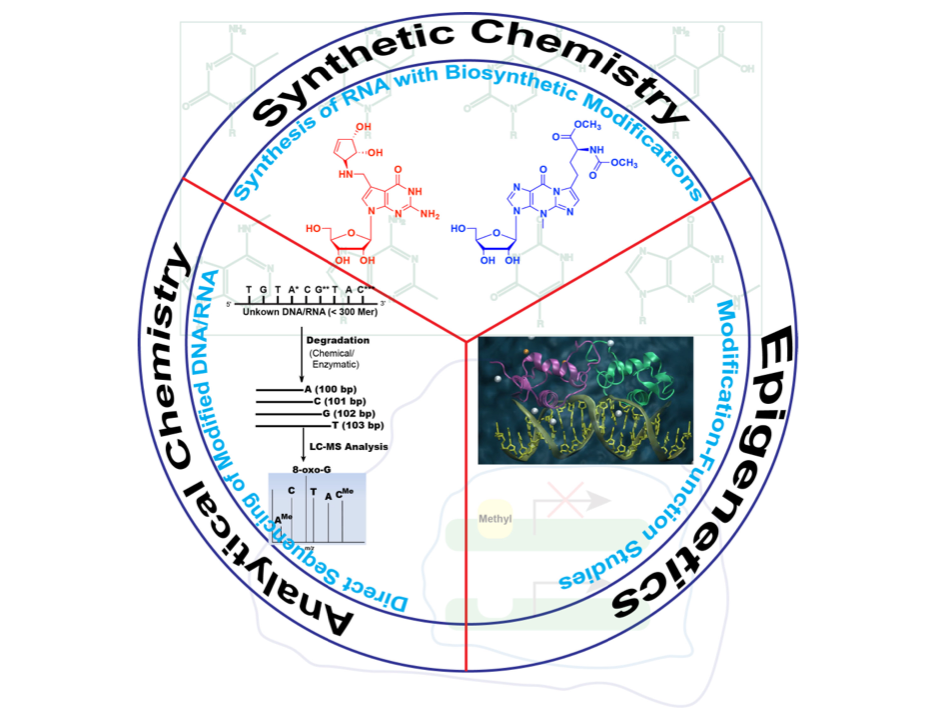
Our interdisciplinary research spans three areas:
- MS-based direct RNA sequencing
- Functional studies of RNA and its modifications
- Synthesizing RNA modifications for functional analysis and therapeutic potential
These synergistic approaches enhance our understanding of RNA functions and applications.
Support the Zhang Lab
Our highly innovative and sustainable research program is supported by both federal and private funding. If you are interested in supporting our work, please feel free to contact Dr. Zhang directly at [email protected].
1400 Washington Avenue
Albany, NY 12222
United States