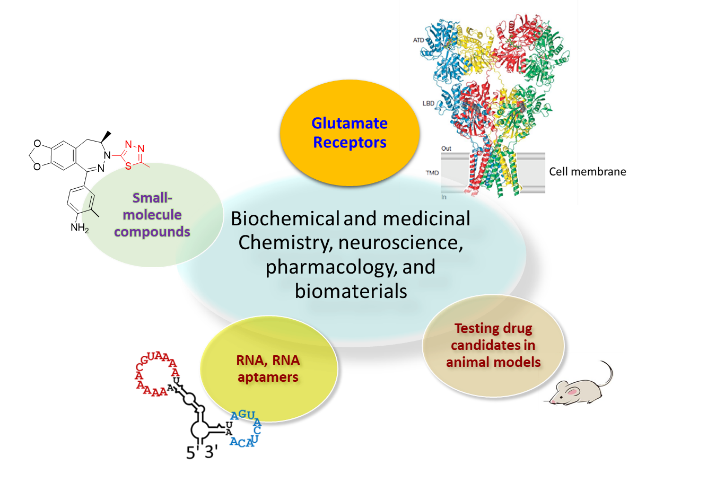
Research

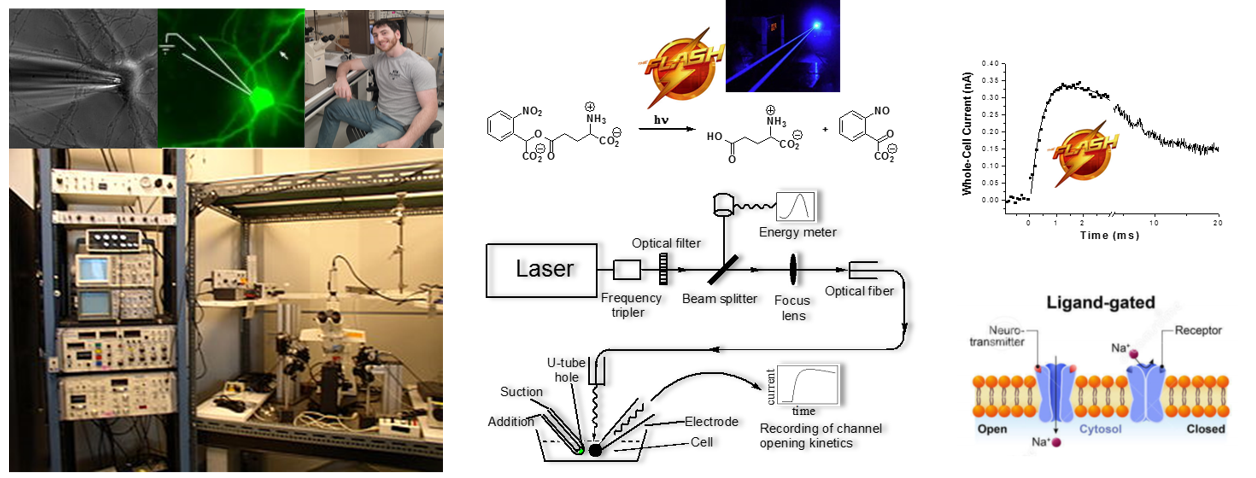
Structure-function relationship of glutamate ion channels
The three-dimensional structure of a protein defines not only its size and shape, but also its function. Understanding the relationship between protein structure and function is one of the foremost challenges in finding ways to regulate the in vivo function of a protein with precision. Projects in this area of our lab are focused on rapid kinetic measurement of the channel-opening rate on the microsecond time scale by using a laser-pulse photolysis technique with “cage glutamate”. These measurements are essential towards a better understanding of the mechanism of action for various subunits/isoforms of glutamate ion channels with both homomeric and heteromeric configurations and also with and without auxiliary subunits, and developing effective means and molecular reagents to more precisely control the function of the receptor activity. Currently, we are focusing on AMPA and kainate receptor subtypes and their isoforms in each subtype.
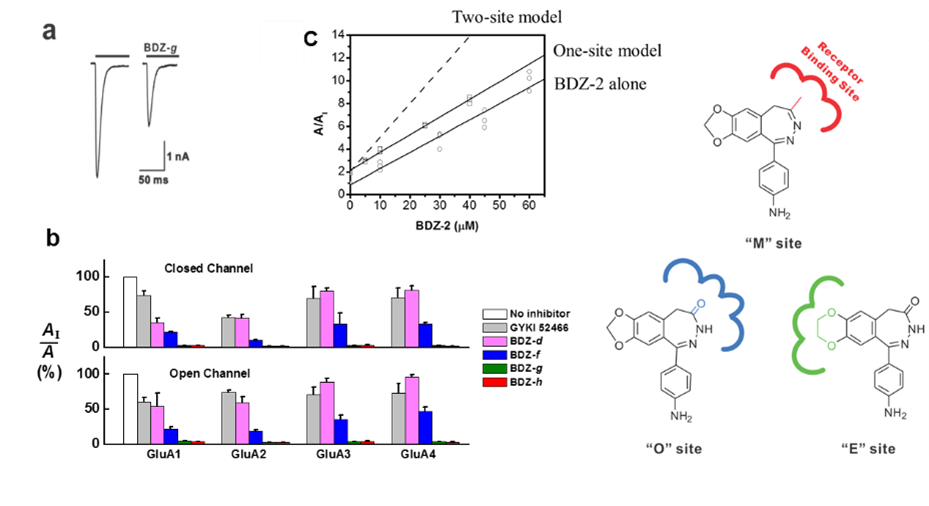
Developing small-molecule, regulatory agents (inhibitors and potentiators) as potential drug candidates
This area focuses on developing small-molecule compounds and understanding the mechanism of action as well as the site of interaction with receptor targets. The goal is to modulate the activity of the target in vivo with high potency and selectivity. These small-molecule compounds, prepared by synthetic chemistry, are potential drug candidates for a number of neurological diseases such as ALS, epilepsy and stroke. Positive modulators are potential drug candidates for cognitive deficiency such as memory deficits in Parkinson’s disease. We also characterize the structure-activity relationship for these compounds.
RNA – Aptamers as potential drug candidates
One of the core components in our research is discovery of RNA molecules as water-soluble, potent and selective antagonists or positive modulators (potentiators) of glutamate ion channels. We use SELEX technique (see the graph here) to isolate these useful aptamers from a large RNA library (~1014 sequences). We also make chemically modified RNA aptamers by replacing 2’-OH group in ribose so that these RNA aptamers are long lasting, and are therefore amenable for in vivo use as drug candidates for CNS diseases. We are also using molecular folding and docking tools, together with mutational analysis, to design RNA aptamers, based on some of the RNA templates discovered through SELEX, so that we can generate RNA aptamers with a more predictable property. Our goal is to make a group of potent, both subtype- and subunit-selective RNA aptamers as a new class of regulatory molecules to control (i.e., inhibit or potentiate) glutamate receptor activity.
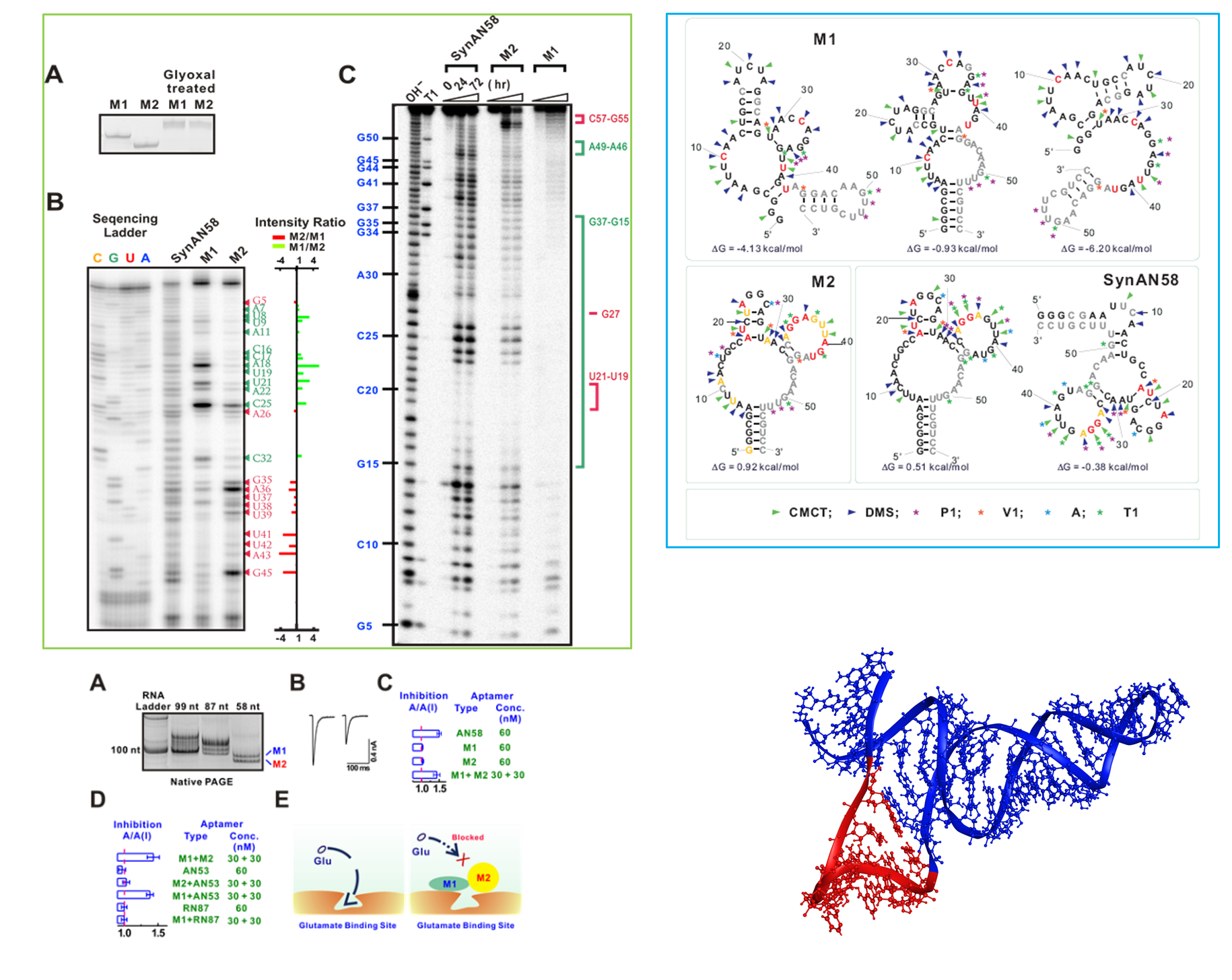
RNA – Folding and function
This area of research focuses on understanding of RNA folding into stable structures for a myriad of functions, such as modulating protein activities as drug candidates. RNA has extensive intramolecular interactions that cause it to fold from a linear molecule with specific sequence into an array of complex structures. RNA folding begins as the transcript is synthesized by RNA polymerase. The formation of RNA structures, such as hairpins, on the nascent transcript can create physical barriers that prevent backtracking and promote certain pathways favor specific tertiary structures with unique functional properties. Here we study the folding of an RNA, which can adopts multiple structures (not reversible, dynamic conformations) with distinct functions from transcription (i.e., copying of a single DNA sequence into RNAs by RNA polymerase).
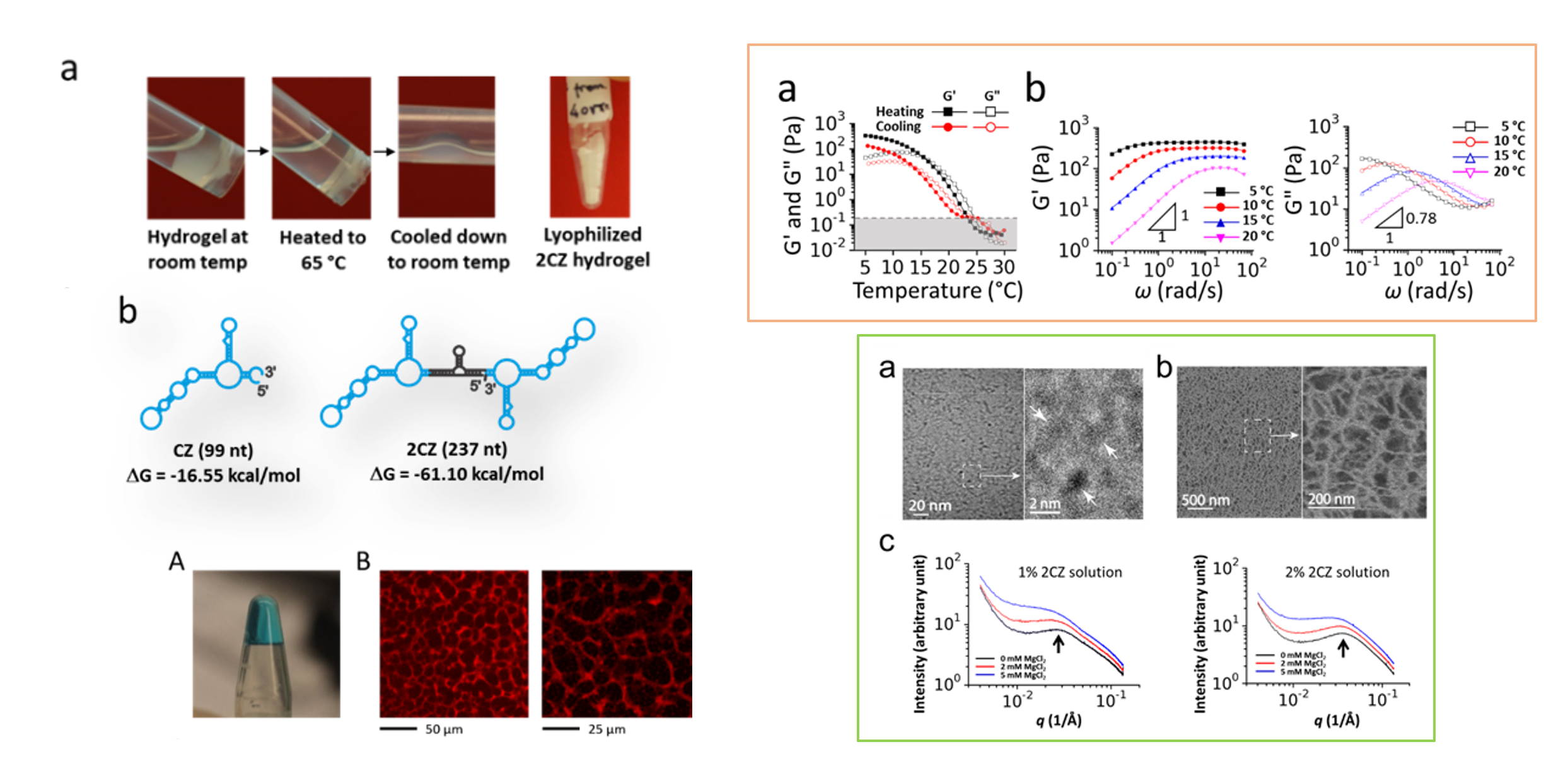
RNA – Hydrogel and biomaterials
Hydrogels are supramolecular assemblies with both solute transport properties like liquids and mechanical properties like elastomers. To date, every type of biomolecules, except RNA, are capable of forming hydrogel. Recently, we have found a unique RNA, which can self-assemble, without any cross-linker or any external support matrix, to form hydrogel. In this case, a single RNA is the monomer, which contains proper recognition domains for intermolecular, non-covalent interactions in order to self-assemble into the hydrogel network structure. Hydrogels formed from biomolecules like RNA are generally biocompatible and can be chemically and biologically decorated and functionalized, making them particularly useful for tissue engineering and drug delivery, etc. We are identifying design modules and making various RNA hydrogels with different mechanical and physical properties. We are testing them using a variety of techniques such as SAXS, cryo-SEM and cryo-TEM.
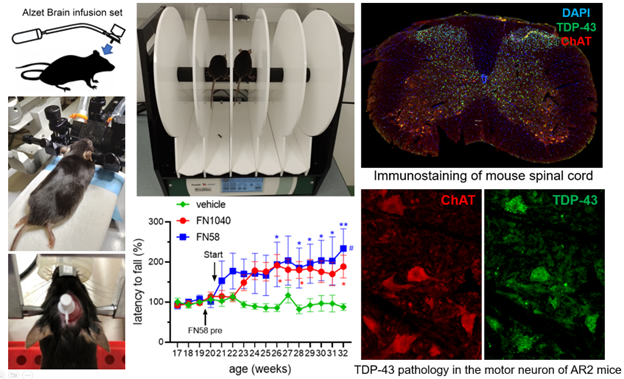
Testing RNA aptamers with animal models of human diseases
Discovery of a drug candidate in terms of its efficacy and its potential toxicological effects requires in vivo study, and animal models are the most practical tools for in vivo, pre-clinical study of a drug candidate before its application in clinical trials. Currently, we are testing a group of RNA aptamers with two transgenic mouse models that mimic familial and sporadic ALS. ALS is a fatal type of motor neuron disease, and currently there is no cure. In one of our studies, for example, we examined the efficacy of an aptamer infused through intraventricular injection on mice with dysregulated AMPA receptor activity. We found that administration of the aptamer rescued both the number and the size of the motor neurons, and the mice performed better in motor function. We are also testing our RNA aptamers with a rat model that mimics neuropathic pain induced by spinal cord injury (SCI). Neuropathic pain caused by a lesion or disease of the somatosensory nervous system is a common chronic pain condition with major impact on quality of life. Most of the available treatments for neuropathic pain have moderate efficacy and present side effects that limit their use. Our preliminary study has shown that administration of RNA aptamers to rats with SCI has generated strong, long-lasting analgesic efficacy without any detectable side effects.
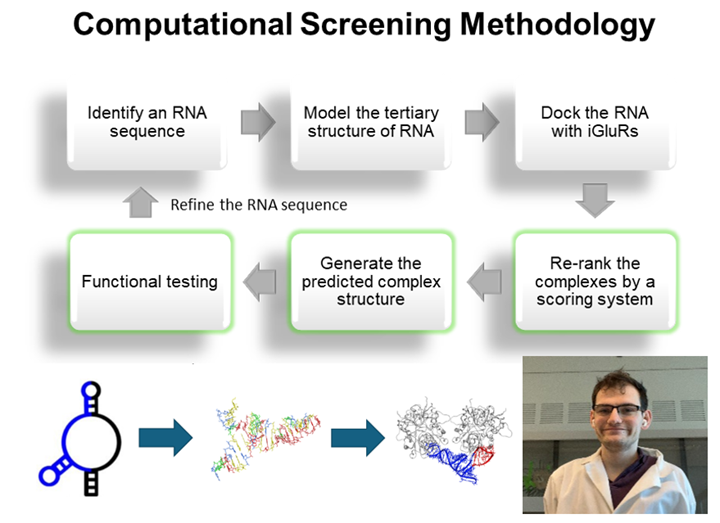
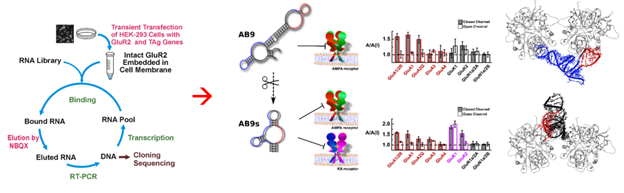
RNA-receptor molecular docking: Using computational approach for discovery of highly selective RNA aptamers
Computational modeling of RNA-glutamate receptor interactions has been a recent addition as a tool for our effort to develop RNA aptamers with high potency and selectivity. This method is especially useful when combined with RNA sequence mutations and then functional testing of the sequence variants with individual glutamate receptor channels using electrophysiology. The diagram to the left shows a flowchart of our approach.